Today, the Montreal Heart Institute (MHI) Cardiovascular Genetics Centre (CGC) is the only centre in Quebec with a unique and exhaustive expertise in the follow-up of individuals that combines clinical expertise and molecular analysis of genetic cardiovascular diseases. The CGC is composed of two sections: the Genetics Clinic and the Molecular Diagnostic Laboratory.
The mission of the Genetics Clinic is to combine innovative strategies for the diagnosis and management of hereditary cardiovascular diseases with applied knowledge in order to promote preventative healthcare.
The mission of the Molecular Diagnostic Laboratory is to offer, on a provincial scale, a molecular diagnosis service for hereditary cardiovascular diseases.
Specifically, the Centre offers:
- A clinical evaluation of individuals with a hereditary cardiovascular disease (suspected or diagnosed)
- Genetic evaluation and genetic counselling for patients and their families
- Molecular genetic analysis for patients and their families
- Comprehensive treatment of patients affected with a hereditary cardiovascular disease.
Laboratory
In addition to the analysis of Single Nucleotide genetic Variations (SNV), small genetic variations (InDel) and Copy Number Variations (CNV), this new service offer now includes a wider range of variants as well as the screening of intronic variants of clinical interest (see list in ‘’Our services’’ section).
In accordance with the principles established by the Ministère de la santé et des services sociaux (MSSS) and the recommendations of the Réseau Québécois de Diagnostic Moléculaire (RQDM), the genes targeted in the various cardiogenetic analysis profiles are approved by a group of RQDM cardiogenetic experts.
The analysis profiles and sequenced genes for familial dyslipidemia are currently being reviewed by a group of experts from the Réseau Québécois de Diagnostic Moléculaire (RQDM). They will be updated in the coming year.
Refer to the Test Request section below for all required documentation for a test submission.
GENERAL PRESENTATION OF THE LABORATORY
All the activities of the MHI's Molecular Diagnostic Laboratory are carried out in Quebec from permanent facilities located on the first floor of the center block of the Montreal Heart Institute.
In order to fulfill its mission and achieve customers needs and expectations, the laboratory relies on a comprehensive Quality Management System. Since 2019, the Molecular Diagnostic Laboratory is a Medical Laboratory accredited by the Standards Council of Canada (SCC) and complies with the requirements of the international standard ISO 15189:2012 and the accreditation conditions established by the SCC.

The Molecular Diagnostic Laboratory participates in many interlaboratory comparison programs of the College of American Pathologists (CAP) and the European Molecular Genetics Quality Network (EMQN). It also carries out the evaluation of its training and professional development programs and ensures personnel takes part in continuing education and skills.
The laboratory is headed by a Clinical Biochemist certified by the Ordre des chimistes du Québec and by the Canadian Academy of Clinical Biochemistry. The members of the team include, among others, a second Clinical Biochemist also certified by the Ordre des chimistes du Québec, four biochemists (M.Sc. Biochemistry) members of the Ordre des chimistes du Québec, two bioinformaticians (M.Sc. bioinformatics), a quality assurance technician, a quality assurance specialist (M.Sc. Biological science), an administrative technician and five medical technologists members of the Ordre professionnel des technologistes médicaux du Québec.
TEST REQUEST
The mandatory documentation and instructions are available below or in the analysis directory (enter code GENA) at the following address:
https://est.omni-assistant.net/icm-ldm/AutoLogin.aspx?User=LDM_ICM_Web
As per RQDM’s recommendations, any request for molecular analysis must be supervised by a health professional specializing in Cardiovascular Genetics and/or Medical Genetics, excepted for the TTR Amyloidosis profile. Please refer patients to the Cardiovascular Genetics Clinic of the Montreal Heart Institute or another Cardiogenetics Clinic/Genetics Clinic in order to benefit from a complete genetic evaluation.
Test Requisition Forms:
Cardiogenetics – NGS (gene panel), deletion/duplication, familial nucleotidic variant search:
Cardiogenetics Test Requisition Form
Familial Dyslipidemia – NGS (gene panel), deletion/duplication, familial nucleotidic variant search:
Familial Dyslipidemia Test Requisition Form
Consent Form
A consent Form is required for every molecular analysis request. Please ensure to include the following document to your Requisition Form.
Note that the MHI's Consent Form as well as any other Consent Form applicable in the Province will be accepted.
OUR SERVICES
One of the most important laboratories of its kind in Canada, the Molecular Diagnostic Laboratory is in operations since 2006 and issues more than 1000 personalized reports per year. It received in 2021 the Supraregional designation for cardiovascular genetic analyses by the Quebec Ministry of Health and Social Services. In close collaboration with the MHI’s Genetics Clinic, the Molecular Diagnostic Laboratory offers genetic analysis of hereditary cardiovascular pathologies in order to meet the following clinical objectives:
To offer a complete cardiovascular molecular genetic analysis service including the management of the specimen by a highly qualified team:
• Sampling, DNA extraction, sequencing analysis (index case and family member) and personalized classification of each identified variant;
• State-of-the-art protocols and devices while maintaining high quality standards;
• Revision of the variants’ classification;
Turnaround Time:
Proband (gene profile by NGS): < 90 calendar days
Priority Proband (gene profile by NGS): < 28 calendar days
Family member (specific variant search): < 28 calendar days
Currently, the Molecular Diagnostics Laboratory offers molecular test profiles for the following genetic cardiovascular diseases:
Inherited cardiomyopathies:
• Hypertrophic cardiomyopathy
• Dilated or arrhythmogenic left ventricle cardiomyopathy
• Arrhythmogenic right ventricular cardiomyopathy
• TTR amyloidosis
Inherited arrhythmias :
• Andersen-Tawil syndrome
• Brugada syndrome
• Long QT syndrome
• Short QT syndrome
• Catecholaminergic polymorphic ventricular tachycardia
• Cardiac conduction defect
• Unexplained sudden cardiac death
Inherited aortopathies :
• Non-syndromic familial aortic aneurysm
• Syndromic aortic aneurysm
• Classic Ehlers-Danlos syndrome
• Vascular Ehlers-Danlos syndrome
• Loeys-Dietz syndrome
• Marfan syndrome
Familial dyslipidemia:
• Familial hyperalphalipoproteinemia
• Familial hyperchylomicronemia
• Sitosterolemia
In addition to the genes listed in the updated queries, the laboratory service offer also includes analysis of these clinically relevant intronic variants:

METHODS
The MHI’s Molecular Diagnostic Laboratory promotes quality. Sequence analyzes are carried out on site by highly qualified personnel in newly equipped laboratories using state-of-the-art equipment.
The analyzes of the Molecular Diagnostic Laboratory of the Montreal Heart Institute are carried out using next-generation sequencing method (NGS). The library preparation is done by capture using personalized probes (Twist Bioscience), according to the Illumina DNA Prep with Enrichment commercial protocol of the Illumina company (not homologated but validated protocol), which allows to minimally target genes involved in cardiomyopathies, arrhythmias and aortopathies, as well as familial dyslipidemia. The coding exons as well as the intron-exon borders (-10bp on each side of the exon) of these genes, in addition to certain specific intronic variants (indicated in the list of genes analyzed, by gene), are then sequenced with a NextSeq2000 sequencer and analyzed using the Illumina’ s Dragen Bio-IT analysis server. A tertiary analysis then annotates the variants corresponding to the genes of the requested profile. Single nucleotide genetic variations (SNV) or small genetic variations (InDel) are detected by this method with an analytical sensitivity of 99.98% and an analytical specificity greater than 99.99%. Copy number variations (CNV) are detected with a minimum analytical sensitivity of 87% and an analytical specificity of 99.96%.
As needed, direct Sanger sequencing can be used to confirm SNVs/InDels and cover low coverage regions. All reportable CNVs are confirmed by a method described in the interpretation section when applicable.
Positioning and isoform information is assembled on the GRCh38/hg38 v0 reference genome.
Some chromosomal regions are duplicated on the genome with at least 90% homology over more than 1000bp. These segmental duplications affect the quality of alignment and consequently sensitivity and analytical specificity of variant detection during bioinformatics analysis. These regions represent less than 0.36% of all the coding regions of our profiles.
The following table summarizes the regions corresponding to segmental duplications among our profiles:
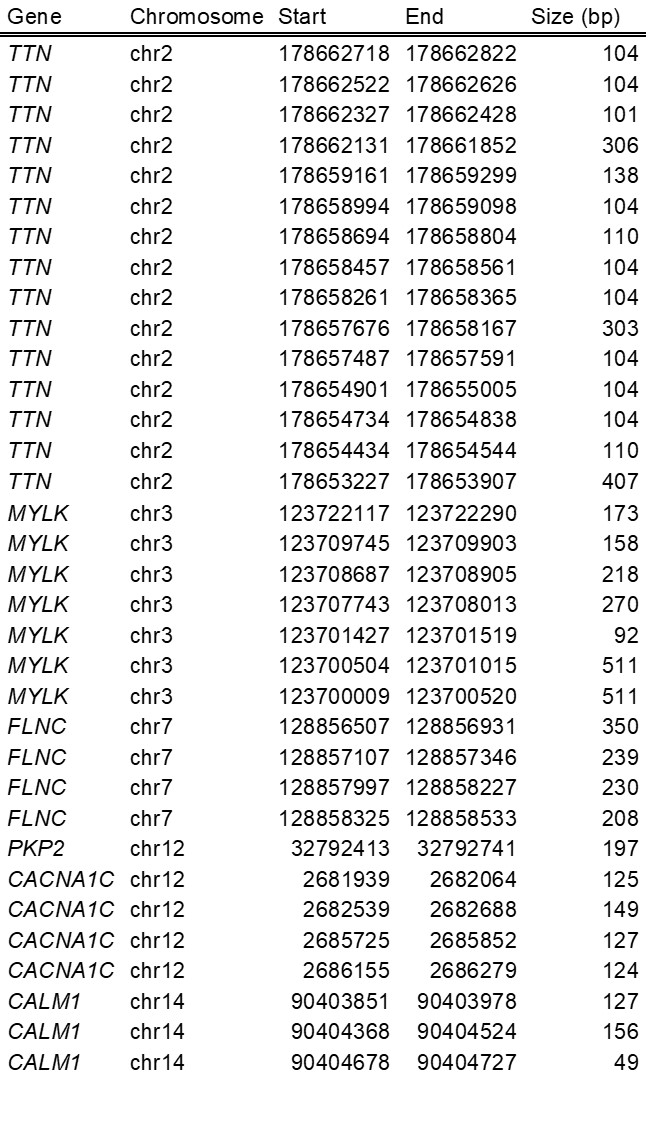
RESULTS INTERPRETATION
All observed variants are reviewed in the literature and annotated using several recognized databases such as ClinVar, gnomAD (genome Aggregation Database), dbSNP) or any other relevant sources of information. An evaluation of the in silico effect of variants on the associated protein is also carried out. The final variant classification is performed according to the criteria of the American College of Medical Genetics and Genomics (ACMG) (PMID: 25741868, 2015).
Although the analysis reports do not mention genetic variants normally present in the population (polymorphisms) or not linked to the disease, they may be reported later whenever future research demonstrates a deleterious effect of these variants. All data is kept secure and accessible for this eventuality.
Since the creation of the laboratory in 2006, the literature review and interpretations have been carried out by a qualified team, which expertise was developed on site and cannot be found anywhere else. The team actively participates in a continuous training program and ensures a sustained technological watch. All results are professionally validated by two certified clinical biochemists.
THE CLINICAL LABORATORY TEAMS
Dr. Julie Amyot, Clinical Biochemist, Ph.D., C.S.P.Q., F.C.A.C.B., Laboratory Director
Dr. Benjamin Neveu, Clinical Biochemist, Ph.D., C.S.P.Q.
Catherine Barahona-Dussault, M.Sc. Biochemist
Guillaume Sylvain-Drolet, M.Sc. Biochemist
Valérie-Anne Codina-Fauteux, M.Sc. Biochemist
Diana Margarita Victoria Morón, M.Sc. Biochemist
Sandra Therrien-Laperrière, M.Sc. Bioinformatician
Valérie Hay, M.Sc. Bioinformatician
Nathalie Lefebvre, Medical Technologist
Marie-Claude Tessier, Medical Technologist
Sadia Ouarezki, Medical Technologist
Nadia Bouzit, Medical Technologist
Roxane Martel, Medical Technologist
Fariza Djerroud, Administrative Technician
Yllmira Darova, Quality Assurance Technician
Valérie Normand, M.Sc. Quality Assurance Specialist
For more informations
For more information, please contact our team, Monday to Friday, 8-17:
Molecular Diagnostic Laboratory
Room C-1760
Montreal Heart Institute
5000 Belanger Street
Montreal (Quebec) H1T 1C8
514-376-3330 ext. 3712
ldm@icm-mhi.org
Please note that administrative personnel can be reached at 514-376-3330 ext. 3066.
Seeking continuous quality improvement, the MHI’s Molecular Diagnostic Laboratory consults its users to ensure that the provided services meet user’s needs and requirements.
As your feedback is important for us, we invite you to fill out this short survey.
Genetic clinic
We offer diagnostic and therapeutic services for the following conditions :
- hypertrophic cardiomyopathy
- familial dilated cardiomyopathy
- arrhythmogenic right ventricle cardiomyopathy
- left ventricle non-compaction cardiomyopathy
- long QT syndrome
- Brugada syndrome (medical information)
- sudden family death
- Marfan syndrome
- Loeys-Dietz syndrome
- Andersen-Tawil syndrome
- Ehlers-Danlos syndrome, Type IV
- family aortic aneurysm
- Polymorphic catecholergic ventricular tachycardia (TVPC)
- amyloidosis
Genetic medical team :
- Dr Mario Talajic
- Dr Patrick Garceau
- Dr Philippe L. L'Allier
- Dr François-Pierre Mongeon
- Dre Lena Rivard
- Dre Anne-Marie Laberge
- Dre Geneviève Giraldeau
- Dr Rafik Tadros
- Dr Philippe Demers
- Dre Sylvia Abadir
- Dre Julia Cadrin-Tourigny
- Dr Jean-Claude Tardif
- Laura Robb, genetic counselor
- Johannie Gagnon, genetic counselor
- Evelyne Naas, clinical nurse
- Karine St-Arnaud, clinical nurse
For more informations
Institut de Cardiologie de Montréal
5000, rue Bélanger
Montréal (Québec)
H1T 1C8
Phone : 514 376-3330 poste 2498
Fax : 514-593-2158
Seeking continuous quality improvement, the MHI’s Molecular Diagnostic Laboratory consults its users to ensure that the provided services meet user’s needs and requirements.
As your feedback is important for us, we invite you to fill out this short survey.
